We use cookies to make your experience better. To comply with the new e-Privacy directive, we need to ask for your consent to set the cookies. Learn more.
Hippo Pathway
Hippo-related products from Covalab
The Hippo signalling pathway has recently emerged as a fundamental regulator of organ size, stem cells function, tissue regeneration and tumor development 1-8. This highly conserved pathway throughout the animal kingdom has been extensively studied on Drosophila melanogaster, and took its name from the major kinase of the cascade, the Hippo kinase.
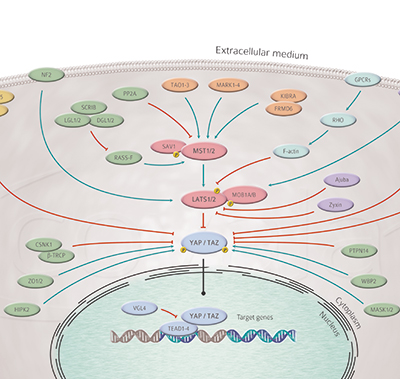
The core “canonical” Hippo pathway is composed of a cascade of kinases, MST1 and 2 being upstream of and activating Lats1 and 2 9-12. Lats kinase, upon activation, phosphorylate the transcriptional co-activators TAZ and YAP which leads to their retention in the cytoplasm. Once Lats are inactivated, the hypophosphorylated Yap/Taz translocate to the nucleus, associate with the Tead family of DNA binding proteins (Tead1 to 4) and activate the transcription of a set of target genes 13-14.
These genes are often linked to promotion of proliferation and inhibition of apoptosis. Several regulators of the pathway have been identified over the years, such as SAV1, MOB1A and MOB1B, FRMD6, the angiomotins, RASSF1, NF2/merlin etc., showing the regulation of Hippo activity to be complex. Indeed, inputs coming from cell surface receptors, adhesion molecules, actin cytoskeleton and metabolic cues were shown to modulate the activity of the pathway 15 for review. This results in a regulation of Hippo activity that is largely tissue-specific. For example, YAP is expressed in many tissues like the liver, intestine 16, heart 17, and pancreas 18, but Yap activity is not always linked to increased proliferation.
The role of the Hippo pathway in cancer is a rapidly growing field. Mutations of specific components of this signalling network have been identified in various cancers. NF2 is inactivated in a rare condition called neurofibromatosis type 2, Lats mutations have been reported in mesothelioma, RASSF1 mutations in lung cancer for example 19-20 for review. Their study holds the promise to identify new therapeutic targets leading to the development of better treatments.
Browse our complete Hippo-related products range or click the image above to display our interactive pathway!
Download Files

Download our dedicated brochure and find what you need among our Hippo-related products!
References:
- 1 - Justice RW et al. Genes Dev. 1995 Mar 1;9(5):534-46
- 2 - Kango-Singh M et al. Development. 2002 Dec;129(24):5719-30
- 3 - Tapon N et al. Cell. 2002 Aug 23;110(4):467-78
- 4 - Harvey KF et al. Cell. 2003 Aug 22;114(4):457-67
- 5 - Jia J et al. Genes Dev. 2003 Oct 15;17(20):2514-9
- 6 - Pantalacci S et al. Nat Cell Biol. 2003 Oct;5(10):921-7
- 7 - Udan RS et al. Nat Cell Biol. 2003 Oct;5(10):914-20
- 8 - Wu S et al. Cell. 2003 Aug 22;114(4):445-56
- 9 - Graves JD et al. EMBO J. 1998 Apr 15;17(8):2224-34
- 10 - Lee KK et al. J Biol Chem. 2001 Jun 1;276(22):19276-85
- 11 - Tao W et al. Nat Genet. 1999 Feb;21(2):177-81
- 12 - Hori T et al. Oncoge- ne. 2000 Jun 22;19(27):3101-9
- 13 - Kanai F et al. EMBO J. 2000 Dec 15;19(24):6778-91
- 14 - Basu S et al. Mol Cell. 2003 Jan;11(1):11-23
- 15 - Johnson R, Halder G. Nat Rev Drug Discov. 2014 Jan;13(1):63-79
- 16 - Zhou D et al. Cancer Cell. 2009 Nov 6;16(5):425-38
- 17 - Heallen T et al. Science. 2011 Apr 22;332(6028):458-61
- 18 - George NM et al. Mol Cell Biol. 2012 Dec;32(24):5116-28
- 19 - Asthagiri AR et al. Lancet. 2009 Jun 6;373(9679):1974-86
- 20 - Hanemann CO. Brain. 2008 Mar;131(Pt 3):606-15.